Events
- Home
- Events
- Development of an atomistic kinetic model of a mAb producing CHO culture
Development of an atomistic kinetic model of a mAb producing CHO culture
Presentation during the 14th European Congress of Chemical Engineering, 17-21 September - Berlin
- by Ypso-Facto
- September 14, 2023
By Alejandro AVILAN GARZON | Ypso-Facto/Université de Lorraine | France
Date: 09-20-2023 , Time: 11:35 am | Topic: G - Tools and Toolkits for Chemical and biochemical reactors
Monoclonal antibodies (mAbs) are complex therapeutic proteins commonly produced via Chinese Hamster Ovary (CHO) cell culture. Like all proteins, mAbs are composed of a sequence of amino acids linked together by peptide bonds. Therefore, from an elemental point of view, they are composed of carbon, hydrogen, oxygen, nitrogen, and sulfur atoms (C.H.O.N.S) that come from substrates such as glucose, glutamine, other essential and non-essential amino acids, as well as other sources present in commercial culture media. The modelling of culture kinetics, especially substrates consumption and cell growth are nearly in all cases simulated using unstructured Monod-type models. However, these models intrinsically verify neither the total mass balance nor the specific mass balance of atoms since, in many cases, only glucose and glutamine are considered as substrates, while, for example, sources of sulfur, an atom present in the main product as well as in the cell, are neglected. The aim of this study is thus to develop an atomistic kinetic model, fundamentally based on the mass balance of C.H.O.N.S.
This innovative approach is based on balances at cell level, with intracellular metabolites accumulation and the coupling of 4 phenomena: I) the transfer of substrates and products across cell membrane, II) the intracellular reactions, III) cell growth and IV) cell death. Concerning the intracellular reactions, instead of using the whole metabolic network, which would involve a high number of species and reactions with complex kinetics expressions and therefore, an important number of parameters to be adjusted, the present model proposes a simplified and novel approach based on the elementary composition (C.H.O.N.S) of each species. Thus, once a substrate crosses the cell membrane, a part of it will accumulate and another part will be splitted into atoms, from which products and other cells will be formed by their assembly. Special attention was taken regarding the mAb production, where instead of using the atomistic approach or another reaction from the literature, it was modeled in an original way as a radical polymerization reaction of an artificial monomer determined from the antibody amino acid composition.
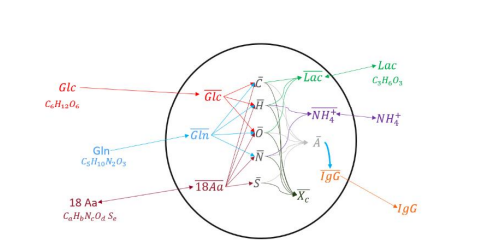
Figure 1. Schematic diagram of the atomistic model.
Internal concentrations are overlined. As for cell growth, instead of using a Monod kinetic growth rate expression, growth was rather related to cell division . In theory, cell mass doubles during the cell cycle, hence the model uses a biomass reaction and the internal biomass accumulation to predict the ending and start of a new cell division cycle. As first approximation, a single cell population was assumed, supposing that all cells grow and behave uniformly. Finally, it was considered that cells could die from a high level of toxic metabolites (lactate and ammonium) and because of substrate limitations.
Figure 1 shows a representation of the model. The developed model was programed using Python and the parameters fitting and model validation were made using data from batch and fed-batch cultures of CHO producing cells carried out in Erlenmeyer flask. Analysis of extracellular and internal concentration of glucose, glutamine, lactate, NH4 + , mAb and lactate dehydrogenase (LDH) were performed using the Gallery multiparametric analyzer (Thermo Fisher Scientific). Amino acids concentration was determined by HPLC-MS. For the intracellular concentrations, a quenching and lysed protocol from literature was used.
The parameters regression was made using the Generalized Differential Evolution 3 (GDE3) genetic algorithm from the python library, Pymoode. The preliminary results show that the model can accurately predict cell growth, substrates consumption and production rates in batch and fed-batch cultures. The experimental determination of intracellular concentrations allowed to validate the model prediction of accumulation of intracellular metabolites. The proposed model presents a greater flexibility in terms of the internal reactions network, which could be replaced with a more or less complex network depending on the desired level of description and the available information.
Recent news

Ypso-Facto is a service company helping industrial firms to develop, optimize and secure their chemical processes and bioprocesses.
Contact Us
-
Address: 19 avenue Foch
54000 NANCY - FRANCE -
LU +352 20 21 39 14
FR +33(0) 355 961 650
- contact@ypso-facto.com