Events
- Home
- Events
- Coupling a macroscopic kinetic model with a flux balance analysis to model a mAb producing CHO culture
Coupling a macroscopic kinetic model with a flux balance analysis to model a mAb producing CHO culture
Presentation during the 14th European Congress of Chemical Engineering, 17-21 September - Berlin
- by Ypso-Facto
- September 14, 2023
Date: 09-19-2023 , Time: 12:25 pm | Topic: H - Digital transformation
Over the last decades, mammalian Chinese Hamster Ovary (CHO) cell culture has become the preferred strategy for industrial therapeutic production of biotherapeutics. This is rooted in their availability to produce, in large quantities and with high quality, complex proteins with post-translational modifications, such as monoclonal antibodies (mAbs) for instance.
Incentives to improve performance, robustness, controllability and automation of the production processes lead to the development of digital twins to simulate and predict the culture behavior, quickly and at low cost. The efficient development of these digital twins are based on biological and physical models, ranging from the microscopic (intracellular) to the macroscopic (extracellular) scale that should be capable of integrating all the necessary process data.
Concerning the microscopic models, developing kinetic models that consider the whole metabolic network is still challenging due to the high number of metabolites and therefore the high number of parameters to be determined. Different alternatives to kinetic models have been proposed to describe the intracellular behavior of cells during the culture like the constraint-based model, Flux Balance Analysis (FBA), that assumes an intracellular steady state (equality of outgoing and incoming fluxes over a short time scale) and determines the intracellular flux distribution using incoming fluxes and an optimality objective (classically maximization of growth rate). However, these steady state models require the input of experimental uptake fluxes limiting their ability to predict dynamic cellular culture behavior. In the present study, the coupling of a macroscopic kinetic model with a flux balance analysis to model the dynamic behavior of a mAb producing CHO cell culture carried out in a bioreactor.
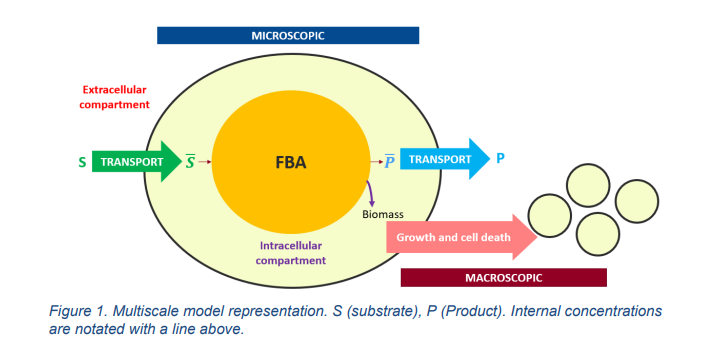
The macroscopic model alone is composed of 4 modules, (i) the transport of substrates and products across cell membrane, (ii) the intracellular reactions, (iii) cell growth and (iv) cell death. The FBA is used to replace the intracellular reactions module, using uptake fluxes determined with the kinetics expression of the macroscopic model depending on external and internal concentrations. The obtained flux distribution as well as the calculated growth rate, allow to determine metabolites internal concentrations that are used as inputs to the other modules of the model (cell growth, cell death and transport across cell membrane). Figure 1 schematizes the coupling of both models.
The developed model was programed using Python and consists of the coupling of an ordinary differential equation system representing all the modules except from the intracellular reaction network, and an FBA module using a genome-scale metabolic model of a producing CHO from literature1 . FBA calculations were made using the COBRApy module from Python. Data from batch and fed-batch cultures of CHO producing cells in Erlenmeyer was used to adjust and validate the model. Analysis of extracellular and intracellular concentration of glucose, glutamine, lactate, NH4+ , mAb and lactate dehydrogenase (LDH) were performed using the Gallery multiparametric analyzer (Thermo Fisher Scientific). Amino acids concentration was determined by HPLC-MS. For the intracellular concentrations, a quenching and lysed protocol from literature was used. The simultaneous resolution of the previously described system results in an original multiscale kinetic model with a detailed description of the intracellular dynamics without the need to include a great number of parameters from complex intracellular kinetics and capable of reproducing the obtained experimental data in terms of cell growth, substrate consumption and production rates.
Recent news

Ypso-Facto is a service company helping industrial firms to develop, optimize and secure their chemical processes and bioprocesses.
Contact Us
-
Address: 19 avenue Foch
54000 NANCY - FRANCE -
LU +352 20 21 39 14
FR +33(0) 355 961 650
- contact@ypso-facto.com